CrossTalkeR Example - Human Myocardial Infarction
Vanessa Klöker
Institute for Computational Genomics, Faculty of Medicine, RWTH Aachen University, Aachen, 52074 GermanyJames S. Nagai
Institute for Computational Genomics, Faculty of Medicine, RWTH Aachen University, Aachen, 52074 GermanyIvan G. Costa
Institute for Computational Genomics, Faculty of Medicine, RWTH Aachen University, Aachen, 52074 GermanySource:
vignettes/MyocardialInfarction.rmd
MyocardialInfarction.rmdCell-Cell Communication Analysis of Human Myelofibrosis scRNA-seq Data Using CrossTalkeR
Here, we demonstrate the usage of CrossTalkeR on the case study of human myocardian infarction snRNA-seq data from the Nature paper “Spatial multi-omic map of human myocardial infarction” (Kuppe et al.). Aim of the study was to build a high-resolution map of human cardiac remodelling after myocardial infarction to provide a better understanding of these remodelling processes. We are focusing here on the conditions “myogenic” (control condition) and “ischemic” (disease condition) and go through the CrossTalkeR results step by step. The original paper already contains CrossTalkeR results and in a later part of this tutorial we will replicate a few of the results.
Data Availability
The data used in this tutorial is available in the following Zenodo
repository: ISMB/ECCB25 -
VT3 tutorial data. We are using the Seurat object
2306_scRNAseq_MOIZ.rds which contains the snRNA-seq data of
the human myocardian infarction study. The object is already
preprocessed and contains the necessary metadata for the CrossTalkeR
analysis.
Since the data set includes all conditions and we intend to use the
python version of LIANA+, we first filter the object to only include the
conditions “myogenic” and “ischemic” and then convert the Seurat object
to an AnnData object. For the conversion we use the
MuDataSeurat package, which allows us to convert Seurat
objects directly to AnnData objects.
# Loading the necessary libraries
library(Seurat)
library(MuDataSeurat)
# Reading the Seurat object
# Note: Please adjust the path to the Seurat object accordingly
complete_heart_data <- readRDS("2306_scRNAseq_MOIZ.rds")
# Filtering the Seurat object to only include the conditions "myogenic" and "ischemic"
filtered_heart_data <- subset(
complete_heart_data,
subset = sampleType %in% c("myogenic", "ischemic")
)
# Saving the filtered Seurat object
saveRDS(filtered_heart_data, "heart_data_object.rds")
# Converting the Seurat object to an AnnData object
MuDataSeurat::WriteH5AD(filtered_heart_data, "heart_data_object.h5ad")Ligand-Receptor Interaction Prediction with LIANA
Before we can run CrossTalkeR, we need the prediction of ligand-receptor interaction for our data set. Due to the large sample size we are using the python version of LIANA for a better performance:
import scanpy as sc
import liana as li
import pandas as pd
import os
out_path = "/out/path/"
data = sc.read_h5ad("heart_data_object.h5ad")
data.raw = data
for i in set(data.obs['sampleType']):
print(i)
lr=li.method.cellphonedb(data[data.obs['sampleType']==i],
groupby='cell_subtype2',
expr_prop=0.1,
verbose=True,
resource_name='consensus',inplace=False)
lr.to_csv(f"{out_path}{i}_lr_liana_consensus.csv")
for i in os.listdir(out_path):
if i.endswith('lr_liana_consensus.csv'):
evfull = pd.read_csv(out_path+i)
evfull = evfull.loc[:,['ligand','receptor_complex','source','target','lr_means','cellphone_pvals']]
evfull['type_gene_A'] = 'Ligand'
evfull['type_gene_B'] = 'Receptor'
evfull['gene_A'] = evfull['ligand']
evfull['gene_B'] = evfull['receptor_complex']
evfull['MeanLR'] = evfull['lr_means']
k = i[0:i.find('_lr_')]
evfull = evfull.loc[list(evfull.cellphone_pvals.to_numpy()<=0.05),:]
evfull = evfull.loc[:, ['source', 'target', 'type_gene_A', 'type_gene_B', 'gene_A', 'gene_B', 'MeanLR']].to_csv(f'{out_path}{k}_lr_ready.csv')Intracellular Communication Analysis with IntraTalker
We can not only analyze the ligand-receptor interactions but also the intracellular communication. For this we use transcription factor activity predictions from decoupler and the IntraTalker package. Similar to usage of LIANA we are using the python version of decoupler:
import scanpy as sc
import decoupler as dc
import pandas as pd
file = "heart_data_object.h5ad"
reg = pd.read_csv("regulon_dorothea_human_AB.csv")
ann_data = sc.read_h5ad(file)
dc.run_ulm(
mat=ann_data,
net=reg,
source='source',
target='target',
weight='weight',
verbose=True,
use_raw=False
)
estimates =ann_data.obsm['ulm_estimate']
estimates.to_csv("/out/path/decoupler_dorotheaAB_results_ulm.csv")To find the transcription factors that show a significant different between the conditions and to connect the transcription factors to upstream receptors and downstream ligands we use the IntraTalker package. First we generate the intracellular communication tables based:
library(Seurat)
library(tibble)
library(dplyr)
library(igraph)
library(stringr)
library(LR2TF)
library(CrossTalkeR)
heart_data_object <- readRDS("heart_data_object.rds")
parameters <- list("out_path" = "/out/path/",
reg = "regulon_dorothea_human_AB.csv",
"organism" = "human",
"celltype" = "cell_subtype2",
"condition" = "sampleType",
"comparison_list" = list(c("ischemic", "myogenic")),
"logfc" = 1,
"pval" = 0.05)
results <- LR2TF::tf_activity_analysis(seuratobject = heart_data_object,
tf_activities = "/out/path/decoupler_dorotheaAB_results_ulm.csv",
arguments_list = parameters)Input Preparation for CrossTalkeR
In the next step, we combine the ligand-receptor and intracellular communication predictions to generate the input for CrossTalkeR:
ischemic_LR <- read.csv("/out/path/ischemic_lr_ready.csv", header = TRUE, row.names = 1)
myogenic_LR <- read.csv("/out/path/myogenic_lr_ready.csv", header = TRUE, row.names = 1)
ischemic_combined <- LR2TF::combine_LR_and_TF_complexes(results@CTR_input_condition$ischemic,
ischemic_LR,
"/out/path/",
"ischemic")
myogenic_combined <- LR2TF::combine_LR_and_TF_complexes(results@CTR_input_condition$myogenic,
myogenic_LR,
"/out/path/",
"myogenic")now the input data for CrossTalkeR is ready and we can run the analysis.
Running CrossTalkeR
The last step is to run CrossTalkeR:
paths <- list('myogenic' = myogenic_combined,
'ischemic' = ischemic_combined)
data <- generate_report(paths,
out_path = "/out/path/",
threshold = 0,
out_file = 'myocardial_infarction.html',
output_fmt = "html_document",
org = "hsa",
comparison = list(c('ischemic', 'myogenic')),
filtered_net = T)
CrossTalkeR Results Ischemic vs Myogenic condition
The second part of this tutorial focuses on the results of the comparative analysis of ischemic versus myogenic conditions.
CCI Results
We start our result analysis with the cell-cell interaction (CCI) analysis of CrossTalkeR and therefore take a look at our CCI network plot:
plot_cci(graph = data@graphs$ischemic_x_myogenic,
colors = data@colors,
plt_name = "ischemic vs myogenic",
coords = data@coords[V(data@graphs$ischemic_x_myogenic)$name,],
emax = NULL,
leg = FALSE,
low = 0,
high = 0,
ignore_alpha = FALSE,
log = FALSE,
efactor = 8,
vfactor = 12,
pg = data@rankings[["ischemic_x_myogenic"]]$Pagerank)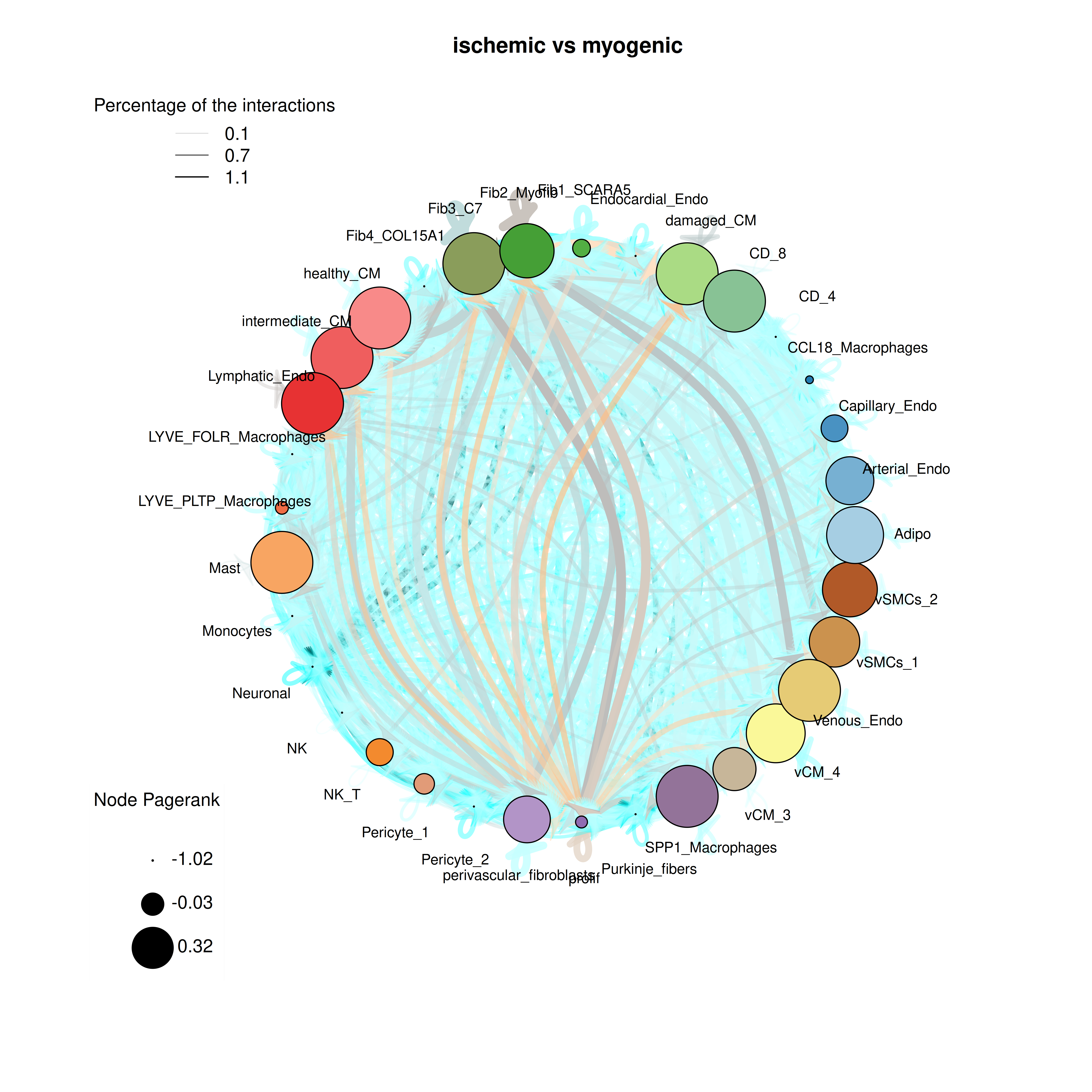
In the ischemic condition, CCIs are generally reduced. However, certain cell types remain notably important, as highlighted by their higher Pagerank scores. These key cell types include cardiomyocyte clusters, two of the four fibroblast clusters, and specific myeloid cells, such as SPP1-expressing macrophages and mast cells. These findings indicate that despite the reduced overall interactions, these cell types are predicted to have a central role in the CCI network of the ischemic condition.
CrossTalkeR also offers a refined CCI network by employing Fisher’s exact test to filter the network based on interaction proportions. We can plot the resulting statistics in a Violon Plot:
EnhancedVolcano(data@stats$ischemic_x_myogenic,
lab = data@stats$ischemic_x_myogenic$columns_name,
x = "lodds",
y = "p",
pCutoff = 0.05)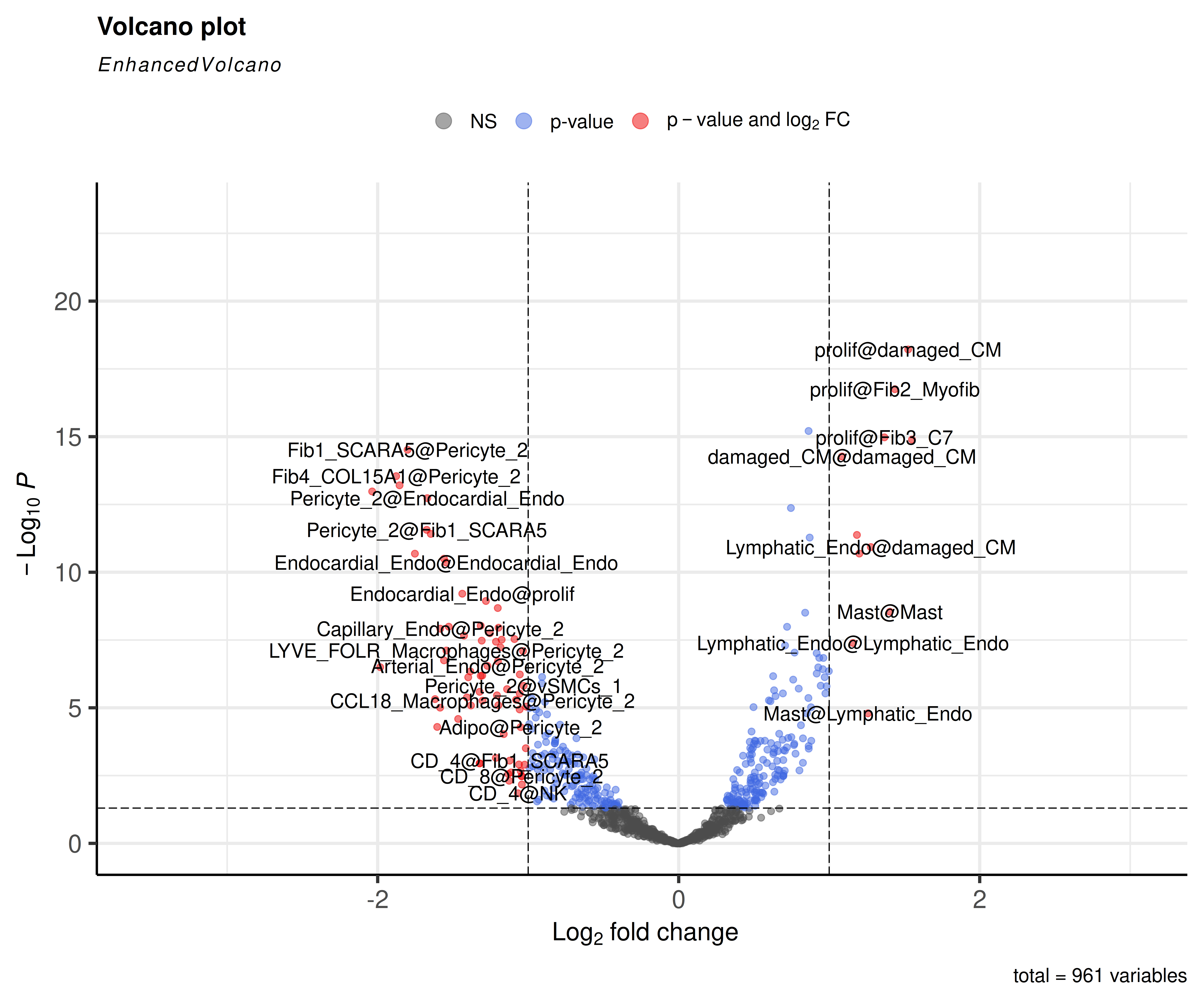
Statistically significant points are labeled with the names of the corresponding edge pairs, with the significance threshold set to a p-value greater than 0.05 and a log2 fold change above one. We see that many of these significant interactions involve high Pagerank cell types, including fibroblast, cardiomyocyte, and myeloid cell clusters. Using this threshold (the default in CrossTalkeR), we can retain only interacting cell pairs with a p-value greater than 0.05 as an example, and replot the CCI network:
plot_cci(graph = data@graphs$ischemic_x_myogenic_filtered,
colors = data@colors,
plt_name = "ischemic vs myogenic filtered",
coords = data@coords[V(data@graphs$ischemic_x_myogenic_filtered)$name,],
emax = NULL,
leg = FALSE,
low = 0,
high = 0,
ignore_alpha = FALSE,
log = FALSE,
efactor = 8,
vfactor = 12,
pg = data@rankings[["ischemic_x_myogenic_filtered"]]$Pagerank)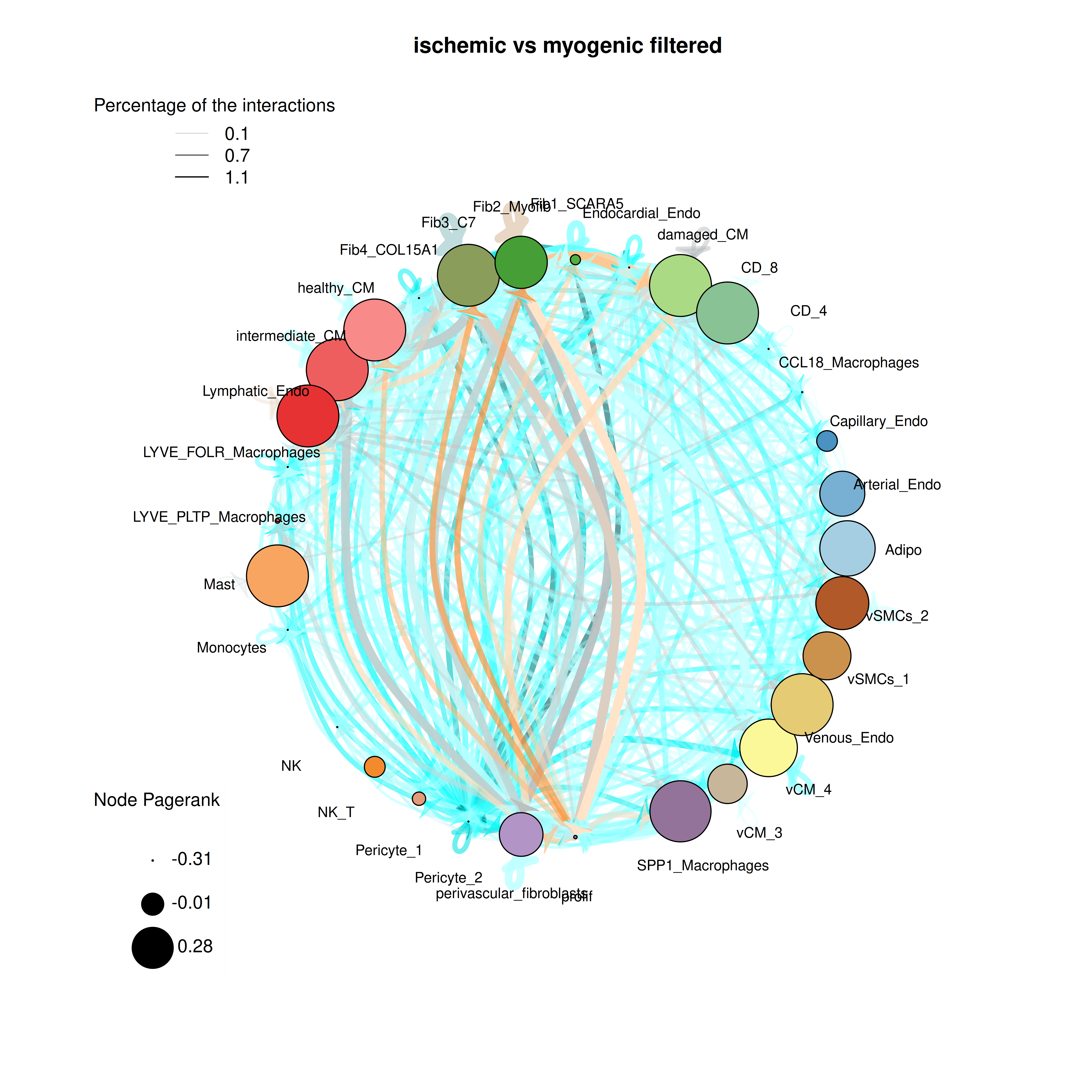
In the filtered CCI plot, positive edges become more visible, though all cell types remain except for the Neuronal and Purkinje_fibers clusters. The Pagerank of the nodes remains largely unchanged.
We can also examine the filtered CCI network with the help of a heatmap:
entries <- names(data@graphs)[grep("_x_",names(data@graphs))]
entries <- entries[grep("filtered",entries)]
for(i in entries){
p1<-plot_Heatmap(graph=data@graphs[[i]],weight = "LRScore")
draw(p1)
}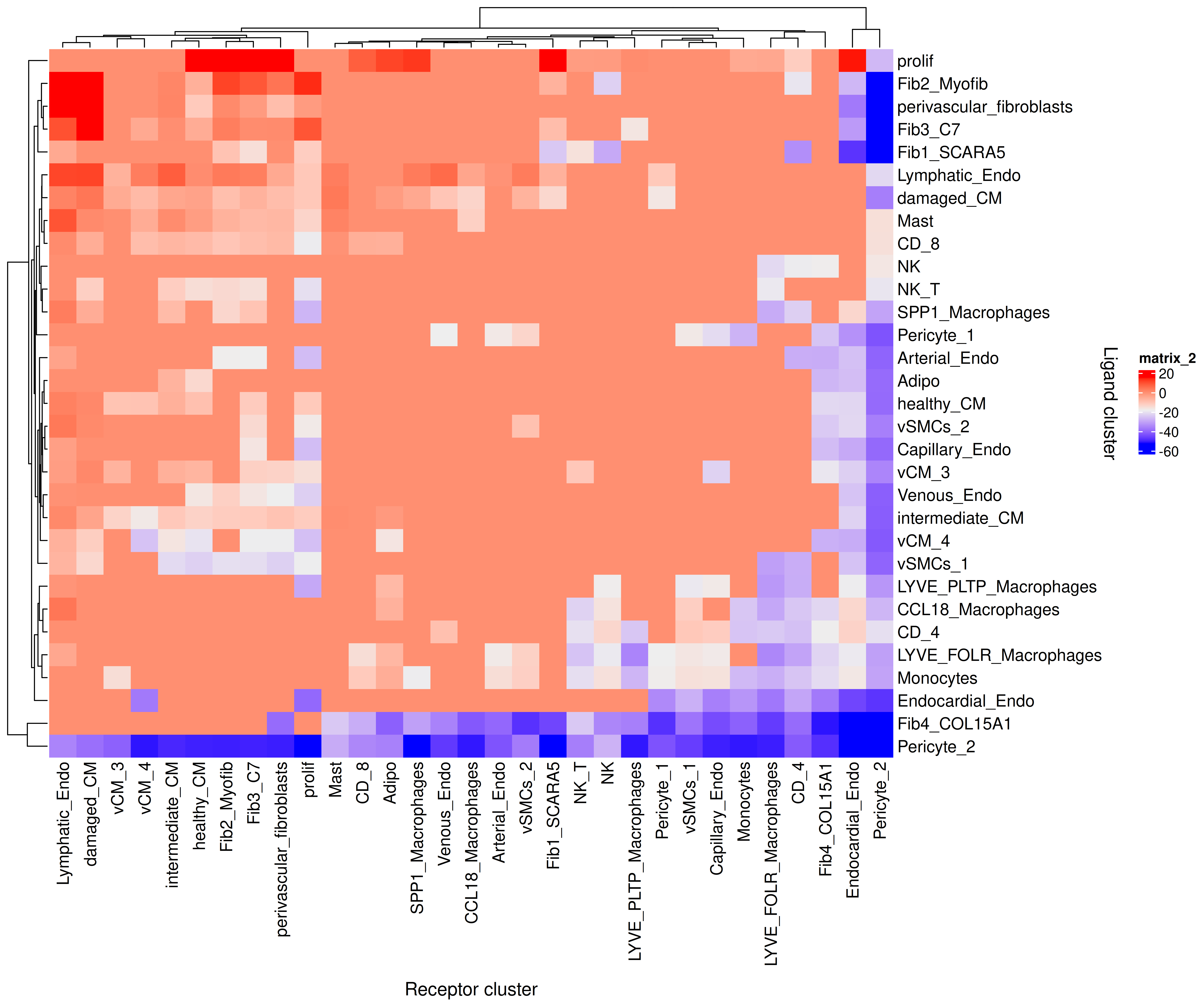
The heatmap provides a more detailed view of the interactions between cell types. The color intensity corresponds to the edge weight in the CCI plot, with red indicating a positive and blue indicating a negative interaction score in the ischemic condition. The sender cell type (Receptor cluster) is on the x axis, and the receiving cell type (Ligand Cluster) on the y axis. We can use the order of the cell types in the heatmap to reorder the nodes in the CCI plot. Here we are sorting the nodes by the order of the sending clusters in the heatmap.
# Extracting the cell cluster order from the Heatmap
row_order <- row_order(p1)
ordered_row_names <- rownames(p1@ht_list[[1]]@matrix)[row_order]
# Generating a new layout template for the CCI plot and define order of the nodes
template <- igraph::make_full_graph(
n = length(ordered_row_names),
directed = TRUE,
loops = TRUE
)
new_circle <- igraph::layout_in_circle(template)
rownames(new_circle) <- ordered_row_names
plot_cci(graph = data@graphs$ischemic_x_myogenic_filtered,
colors = data@colors,
plt_name = 'ischemic vs myogenic filtered',
coords = new_circle[V(data@graphs$ischemic_x_myogenic_filtered)$name,],
emax = NULL,
leg = FALSE,
low = 0,
high = 0,
ignore_alpha = FALSE,
log = FALSE,
efactor = 8,
vfactor = 12,
pg = data@rankings[["ischemic_x_myogenic_filtered"]]$Pagerank,
node_label_position = 1.3)
To confirm the top cell types identified by Pagerank, we can examine their Pagerank scores in more detail using a bar plot:
plot_bar_rankings(data, "ischemic_x_myogenic_filtered", "Pagerank", type = NULL, filter_sign = NULL)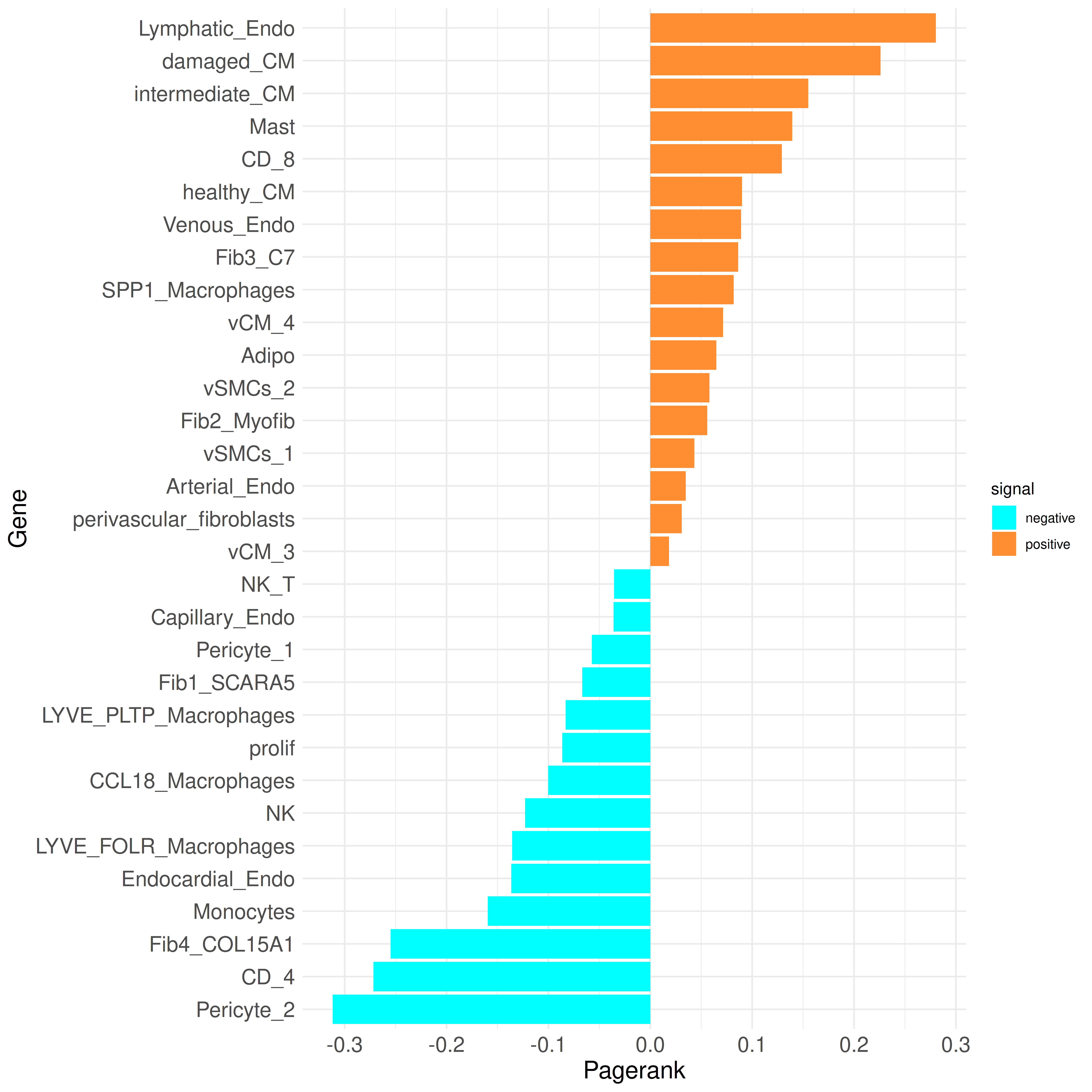
We can further take a look at the Influencer and Listener ranking to observe which cell types are more sending or more receiving:
plot_bar_rankings(data, "ischemic_x_myogenic_filtered", "Influencer", type = NULL, filter_sign = NULL)
plot_bar_rankings(data, "ischemic_x_myogenic_filtered", "Listener", type = NULL, filter_sign = NULL)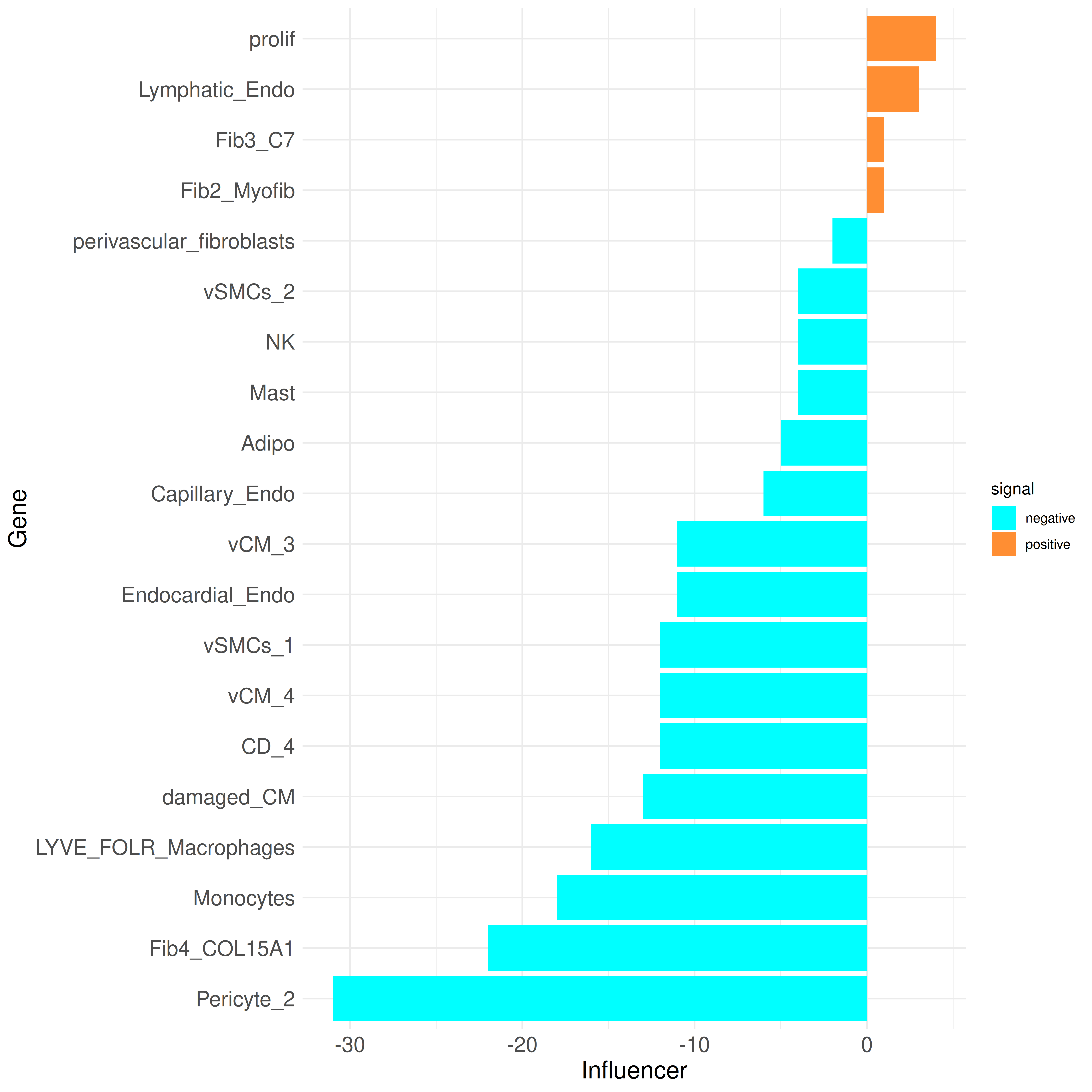
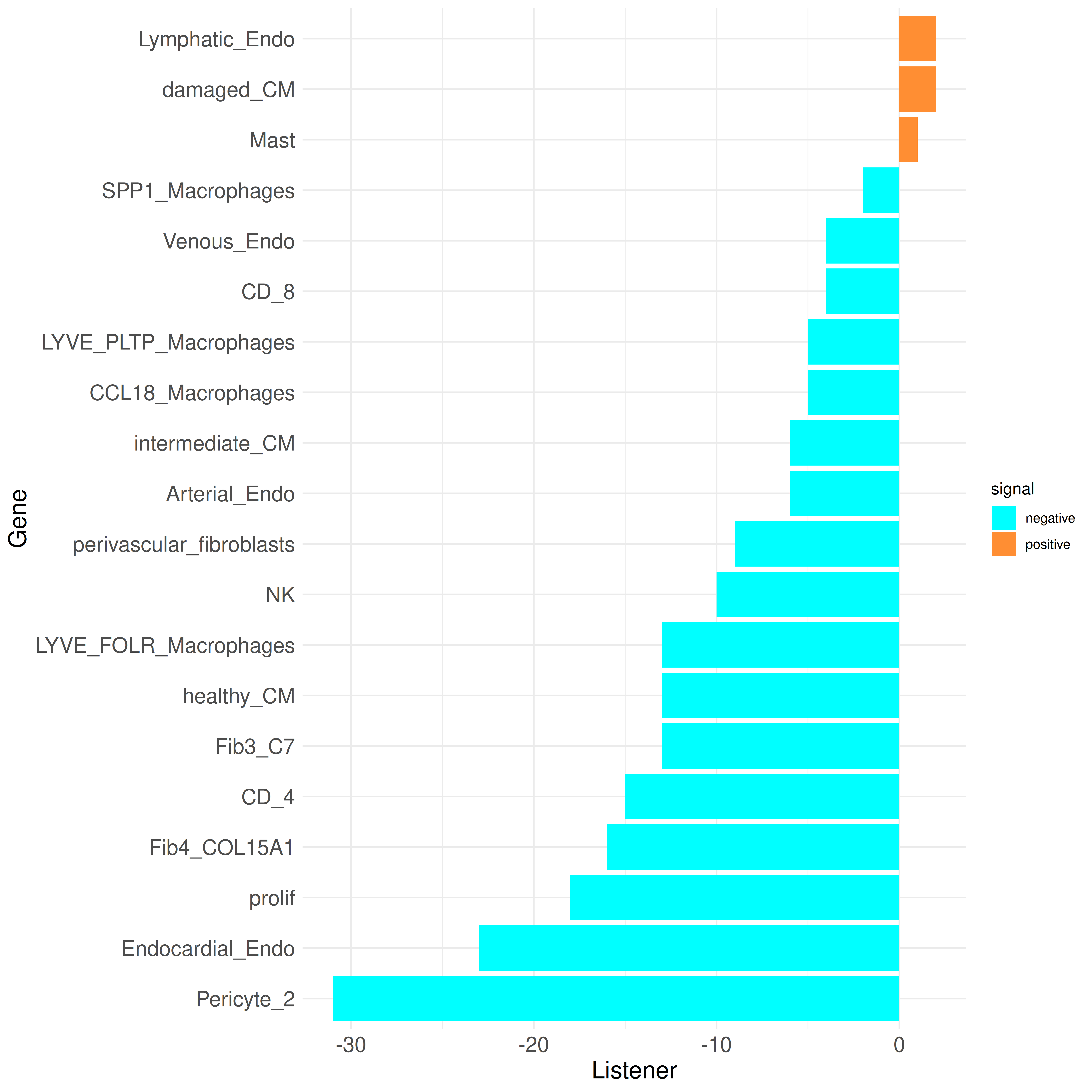
These plots highlight that nearly all cell types are predicted to communicate less in the ischemic condition compared to the myogenic condition. Exceptions include the Lymphatic_Endo and Fib3_C7 clusters, which exhibit a higher Influencer Score in the ischemic condition (indicating more outgoing signals). Additionally, the Lymphatic_Endo, damaged_CM, and Mast clusters show a higher Listener Score, indicating more incoming signals in ischemia.
Most of the observations at the CCI level are consistent with the findings in the paper. For instance, fibrotic structures are more prominent in the ischemic condition, which is also reflected in the Pagerank scores, as at least two out of the four fibroblast clusters have a higher Pagerank in the ischemic condition.
CGI Results
Next, we shift to the gene-cell interaction (GCI) level. Similar to the CCI level, we can examine the network’s topological rankings. We begin by reviewing the top Pagerank results, which identify genes within a specific cell type that are important in the network’s signaling. For now, we will focus exclusively on ligand-receptor (LR) interactions.
plot_bar_rankings(data, "ischemic_x_myogenic_ggi", "Pagerank", type = "LR", filter_sign = NULL, mode = "cgi")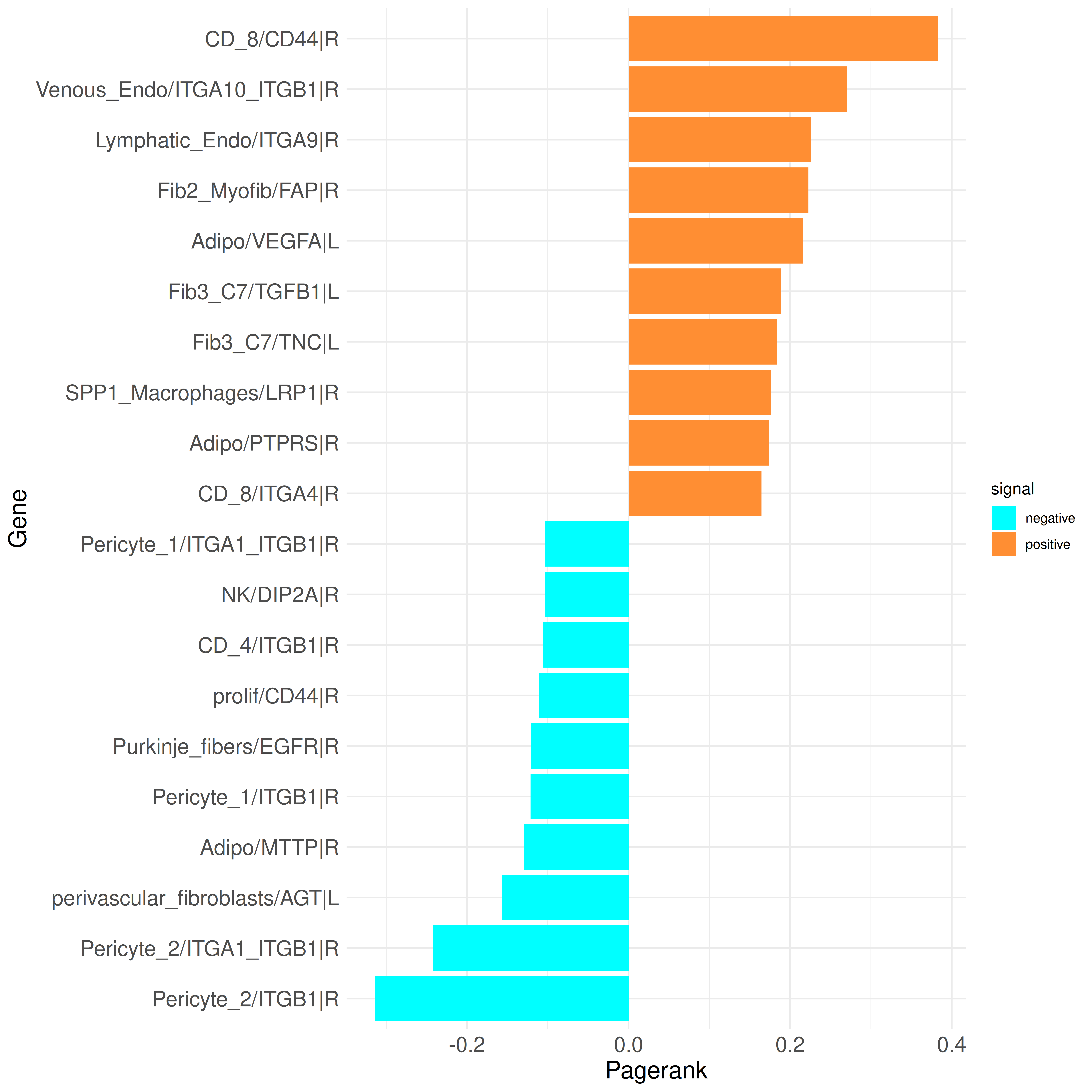
In the top genes, we find both ligands (L) and receptors (R). Many of these genes are involved in immune system processes, such as integrin receptors (e.g., ITGA10_ITGB1, ITGA9, ITGB1, and others). These are predicted to play a larger role in certain clusters (e.g., Venous_Endo, Lymphatic_Endo, SPP1_macrophages) or a smaller role in others (e.g., Pericyte_1, CD_4, Pericyte_2). Additionally, the TGFB1 ligand is among the top genes in the damaged_CM cluster.
To gain a clearer understanding of outgoing and incoming signals, we examine the ligands with the top Influencer scores and the receptors with the top Listener scores:
plot_bar_rankings(data, "ischemic_x_myogenic_ggi", "Influencer", type = "L", filter_sign = NULL, mode = "cgi")
plot_bar_rankings(data, "ischemic_x_myogenic_ggi", "Listener", type = "R", filter_sign = NULL, mode = "cgi")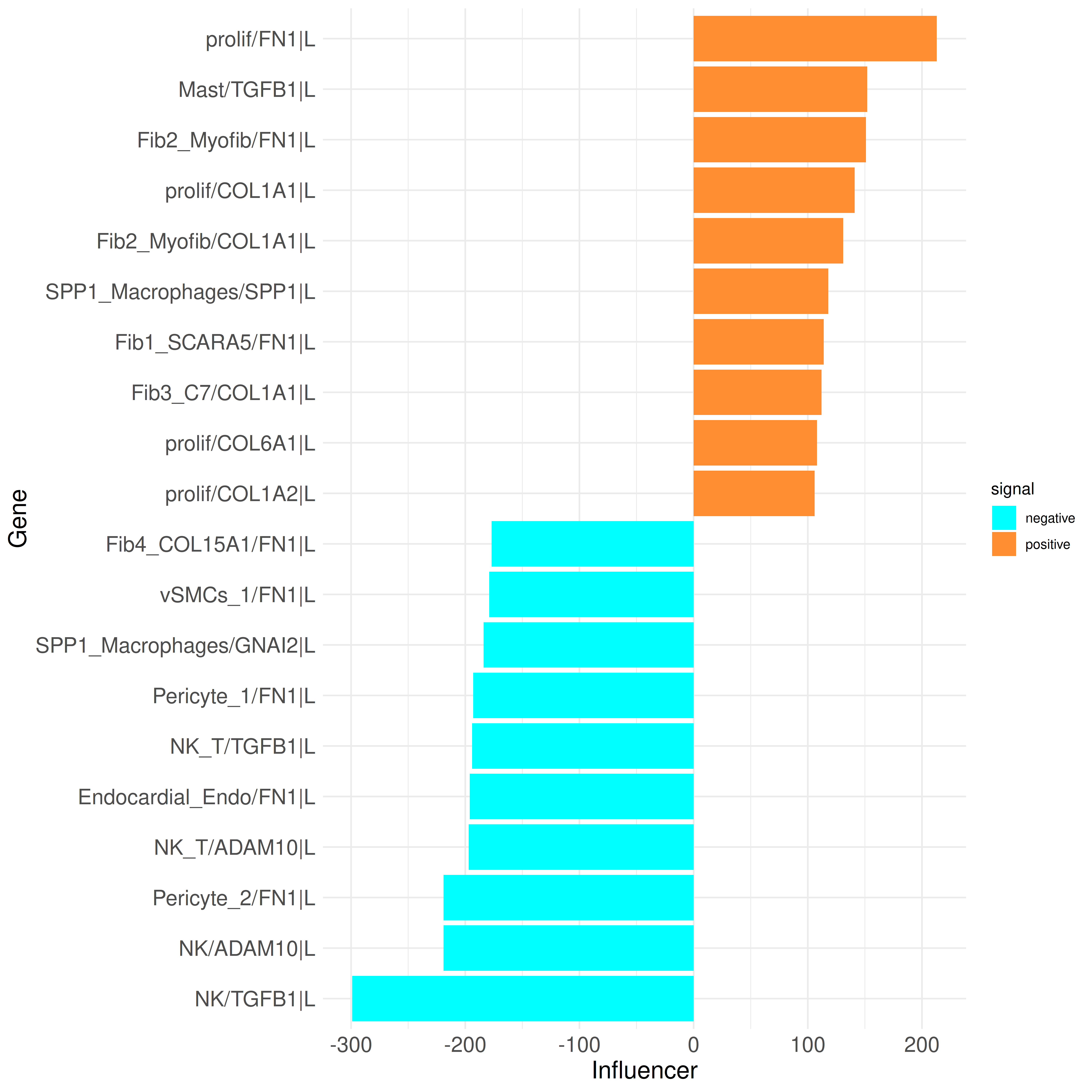
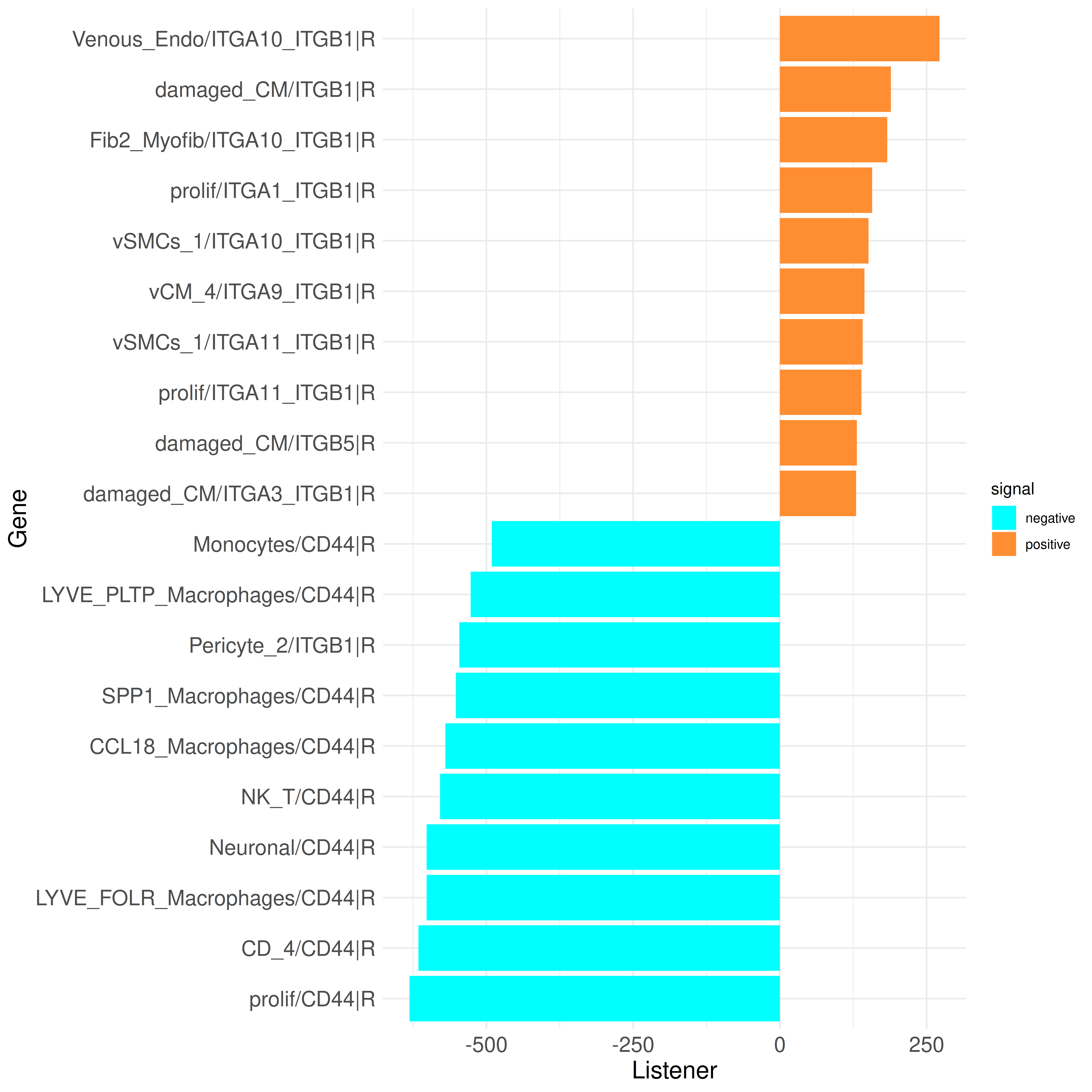
The list of top influencers mainly includes genes important in wound healing, fibrosis, and immune response (e.g., TGFB1, collagens, etc.). The receptors are primarily integrin receptors and subunits and CD44, which play key roles in immune response and tissue repair processes. These results, which have already been described in the paper, further support these findings.
Next, we create the Sankey plots for the ischemic vs. myogenic comparisons, as shown in extended figures 12d and e in the original paper. It is important to note that these plots may differ slightly, as we are using newer versions of the LIANA package and, for simplicity, we are considering fewer samples in this tutorial.
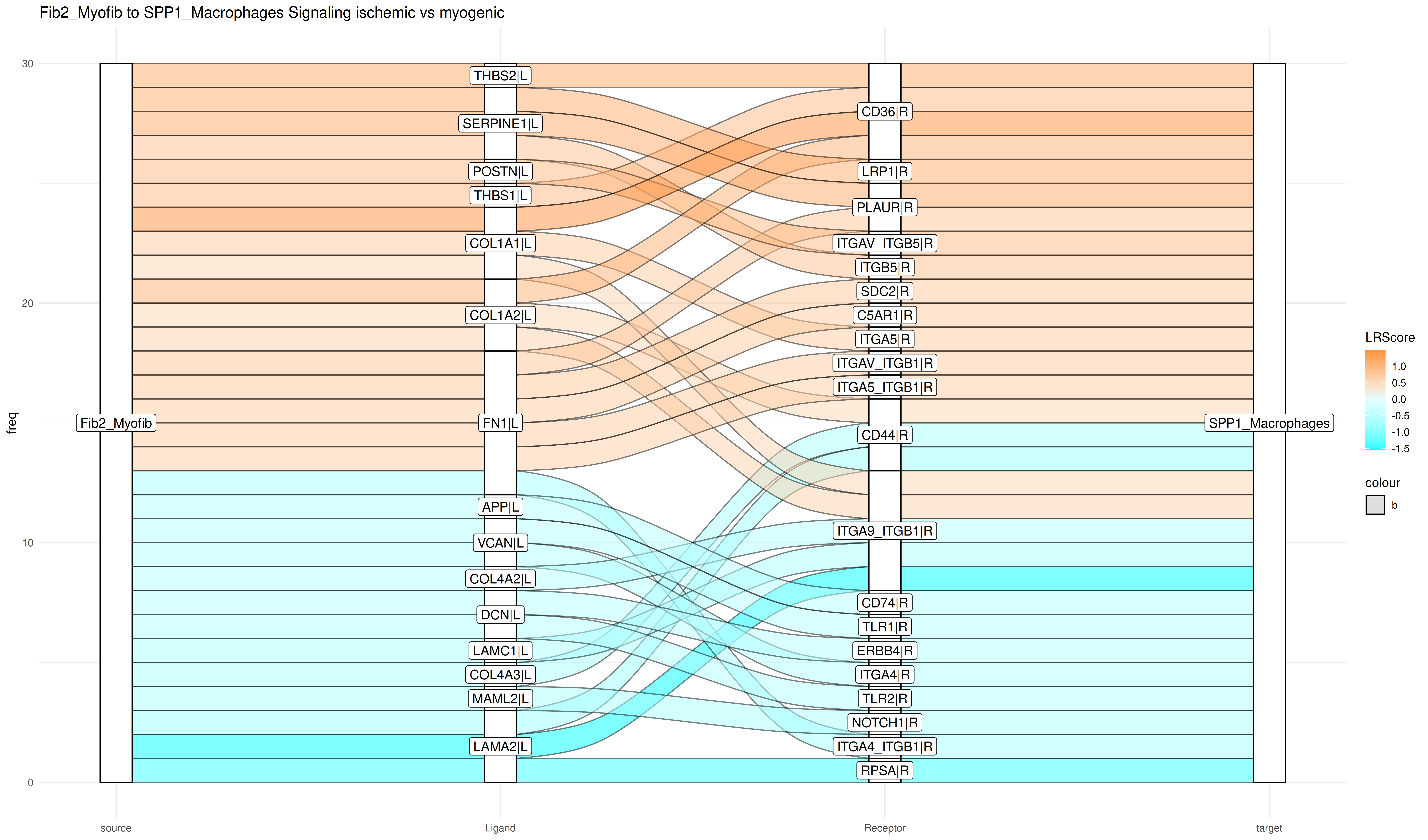
We begin with the Sankey plots showing signaling between the Fib2 and SPP1_Macrophages clusters (in both directions):
plot_sankey(data@tables$ischemic_x_myogenic,
target = NULL,
ligand_cluster = "Fib2_Myofib",
receptor_cluster = "SPP1_Macrophages",
plt_name = "Fib2_Myofib to SPP1_Macrophages Signaling ischemic vs myogenic",
threshold = 30,
tfflag = FALSE
)
plot_sankey(data@tables$ischemic_x_myogenic,
target = NULL,
ligand_cluster = "SPP1_Macrophages",
receptor_cluster = "Fib2_Myofib",
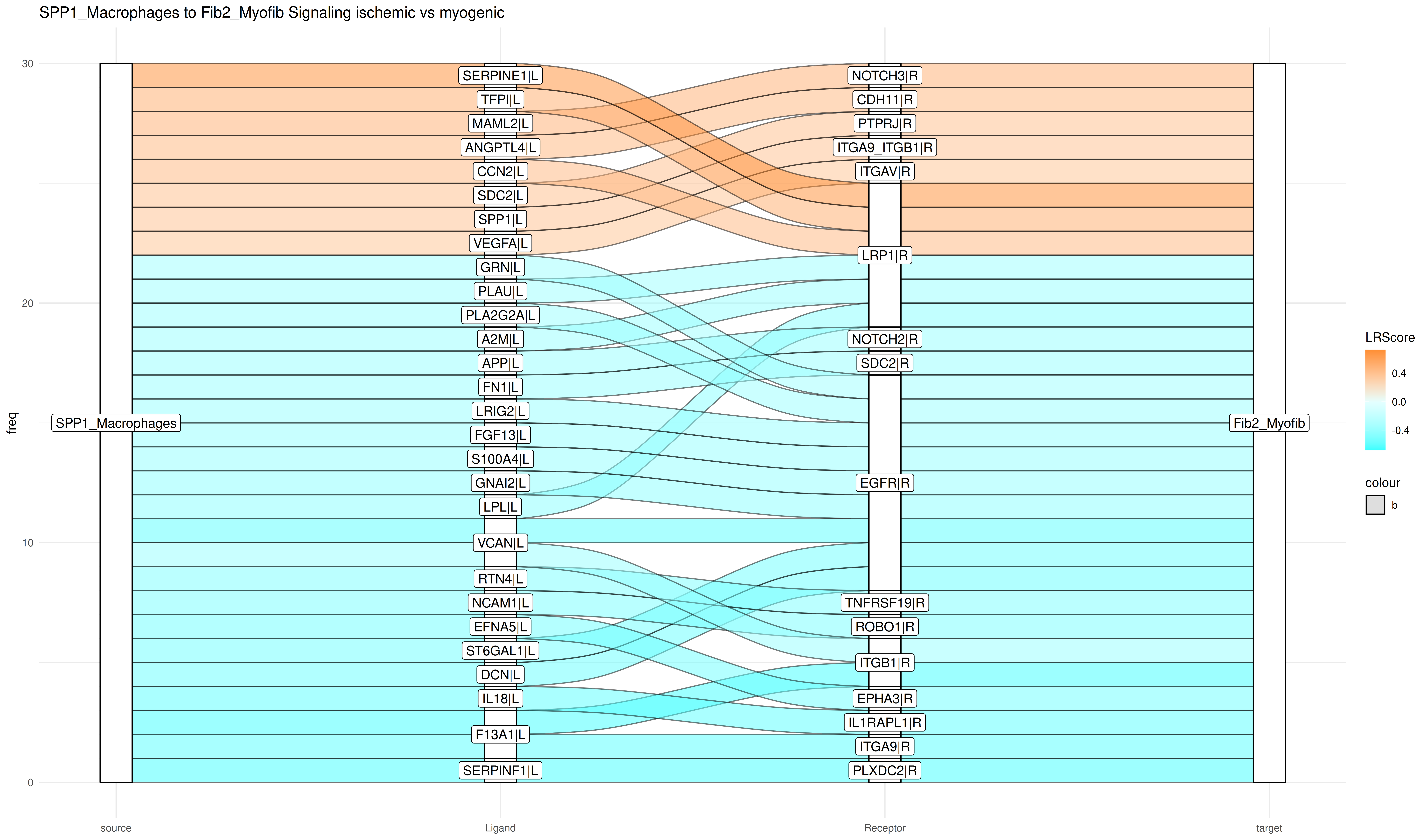
plt_name = "SPP1_Macrophages to Fib2_Myofib Signaling ischemic vs myogenic",
threshold = 30,
tfflag = FALSE
)
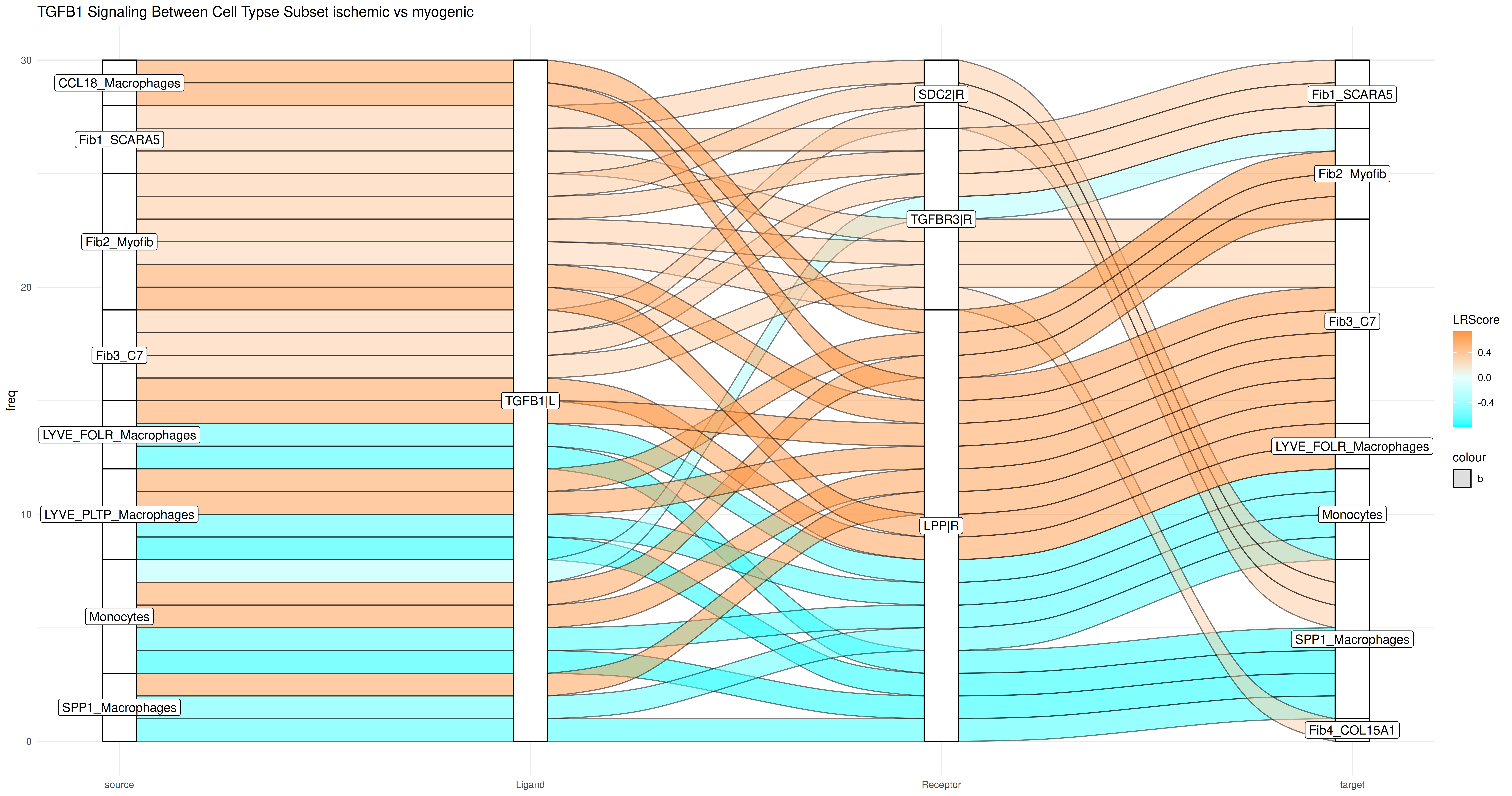
Additionally, a Sankey plot was created for a subset of cell types, displaying only interactions involving the TGFB1 ligand:
subset <- c("LYVE_FOLR_Macrophages",
"LYVE_PLTP_Macrophages",
"CCL18_Macrophages",
"SPP1_Macrophages",
"Monocytes",
"Fib4_COL15A1",
"Fib3_C7",
"Fib2_Myofib",
"Fib1_SCARA5")
plot_sankey(data@tables$ischemic_x_myogenic,
target = "TGFB1|L",
ligand_cluster = subset,
receptor_cluster = subset,
plt_name = "TGFB1 Signaling Between Cell Type Subset ischemic vs myogenic",
threshold = 30,
tfflag = FALSE)