load data
mca.spleen
#> Loading required package: SeuratObject
#> An object of class Seurat
#> 39855 features across 1970 samples within 1 assay
#> Active assay: RNA (39855 features, 0 variable features)
Idents(mca.spleen) <- "anno"
ids <- c("T cell(Spleen)", "Macrophage(Spleen)", "NK cell(Spleen)", "Marginal zone B cell(Spleen)")
mca.spleen@meta.data$annotation_simplified <- mca.spleen@meta.data$annoSimplify the cell annotation
ids <- c("T cell(Spleen)", "Macrophage(Spleen)", "NK cell(Spleen)", "Marginal zone B cell(Spleen)")
mca.spleen@meta.data$annotation_simplified <- ifelse(mca.spleen@meta.data$annotation_simplified %in% ids, mca.spleen@meta.data$annotation_simplified, "Other")
mca.spleen@meta.data$annotation_simplified[mca.spleen@meta.data$annotation_simplified == "T cell(Spleen)"] <- "T Cell"
mca.spleen@meta.data$annotation_simplified[mca.spleen@meta.data$annotation_simplified == "Marginal zone B cell(Spleen)"] <- "B Cell"
mca.spleen@meta.data$annotation_simplified[mca.spleen@meta.data$annotation_simplified == "NK cell(Spleen)"] <- "NK Cell"
mca.spleen@meta.data$annotation_simplified[mca.spleen@meta.data$annotation_simplified == "Macrophage(Spleen)"] <- "Macro"
Idents(mca.spleen) <- "annotation_simplified"Markers identification for NK cell
To run sc2marker you need to execute the following command, providing a clustered single cell data sets (as Seurat object), the cell type of interest and the antibody databased (IHC, ICC or Flow).
nk.markers <- Detect_single_marker(mca.spleen, id = "NK Cell", category = "Flow", org = "mouse", min.pct = 0.1)
get_antibody(nk.markers, org = "mouse")
# Ridge plot of top 9 markers of NK cells
plot_ridge(mca.spleen, id = "NK Cell", genes = nk.markers[1:9,]$gene, ncol = 3, assay = "RNA", aggr.other = F)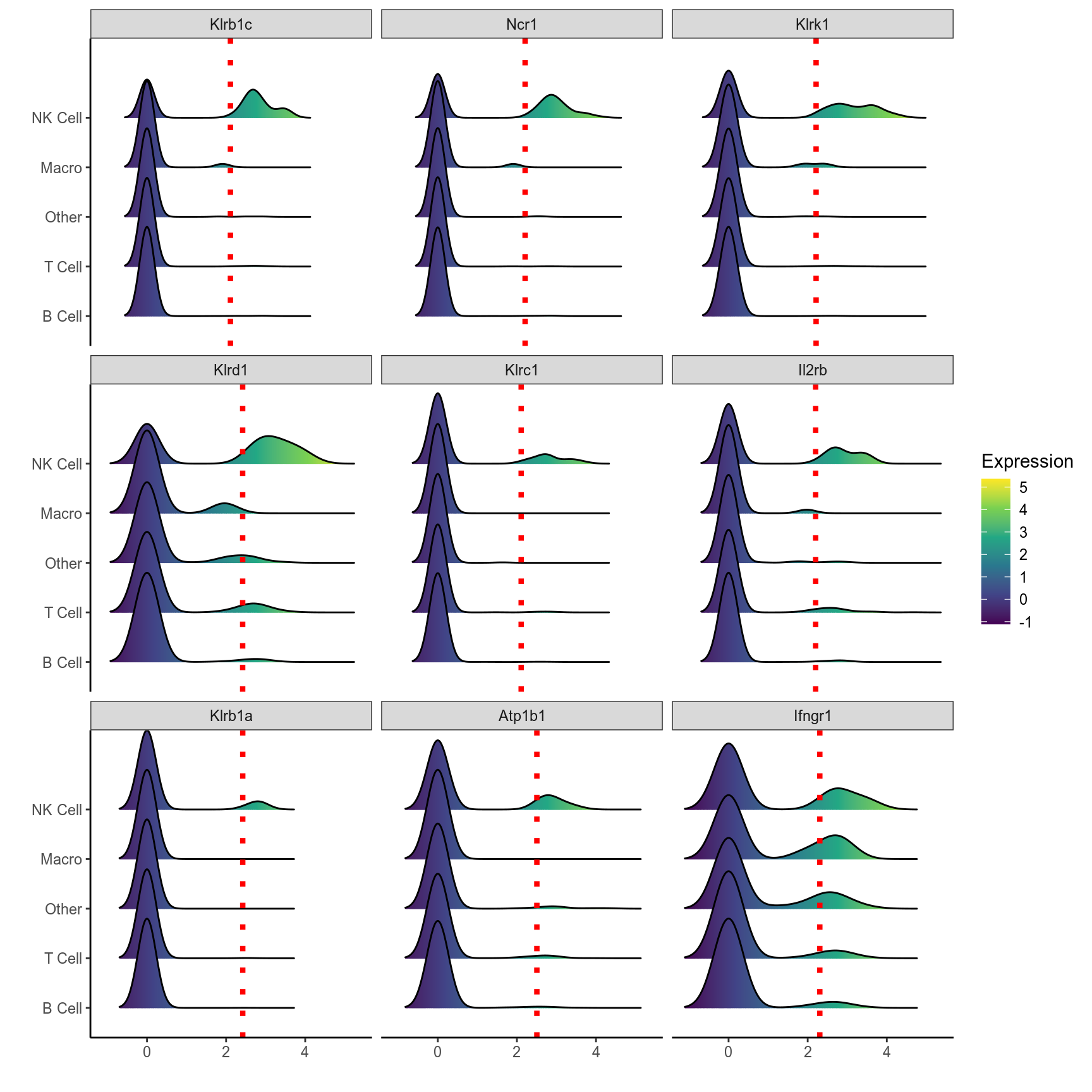
Markers identification for B cell
b.markers <- Detect_single_marker(mca.spleen, id = "B Cell", category = "Flow", org = "mouse", min.pct = 0.1)
get_antibody(b.markers, org = "mouse")
# Ridge plot of top 9 markers of B cells
plot_ridge(mca.spleen, id = "B Cell", genes = b.markers[1:9,]$gene, ncol = 3, assay = "RNA", aggr.other = F)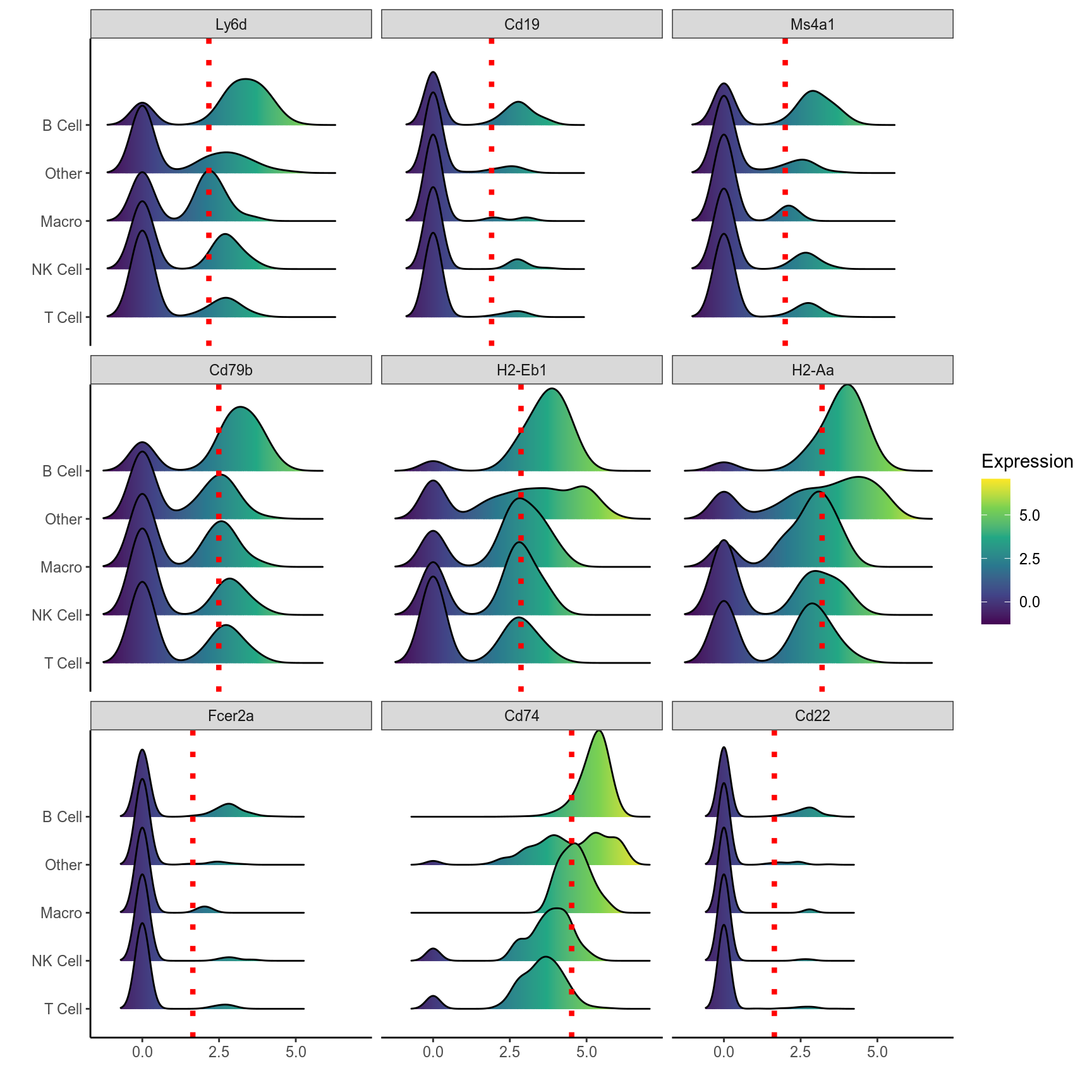
Markers identification for T cell
t.markers <- Detect_single_marker(mca.spleen, id = "T Cell", category = "Flow", org = "mouse", min.pct = 0.1)
get_antibody(t.markers, org = "mouse")
# Ridge plot of top 9 markers of B cells
plot_ridge(mca.spleen, id = "T Cell", genes = t.markers[1:9,]$gene, ncol = 3, assay = "RNA", aggr.other = F)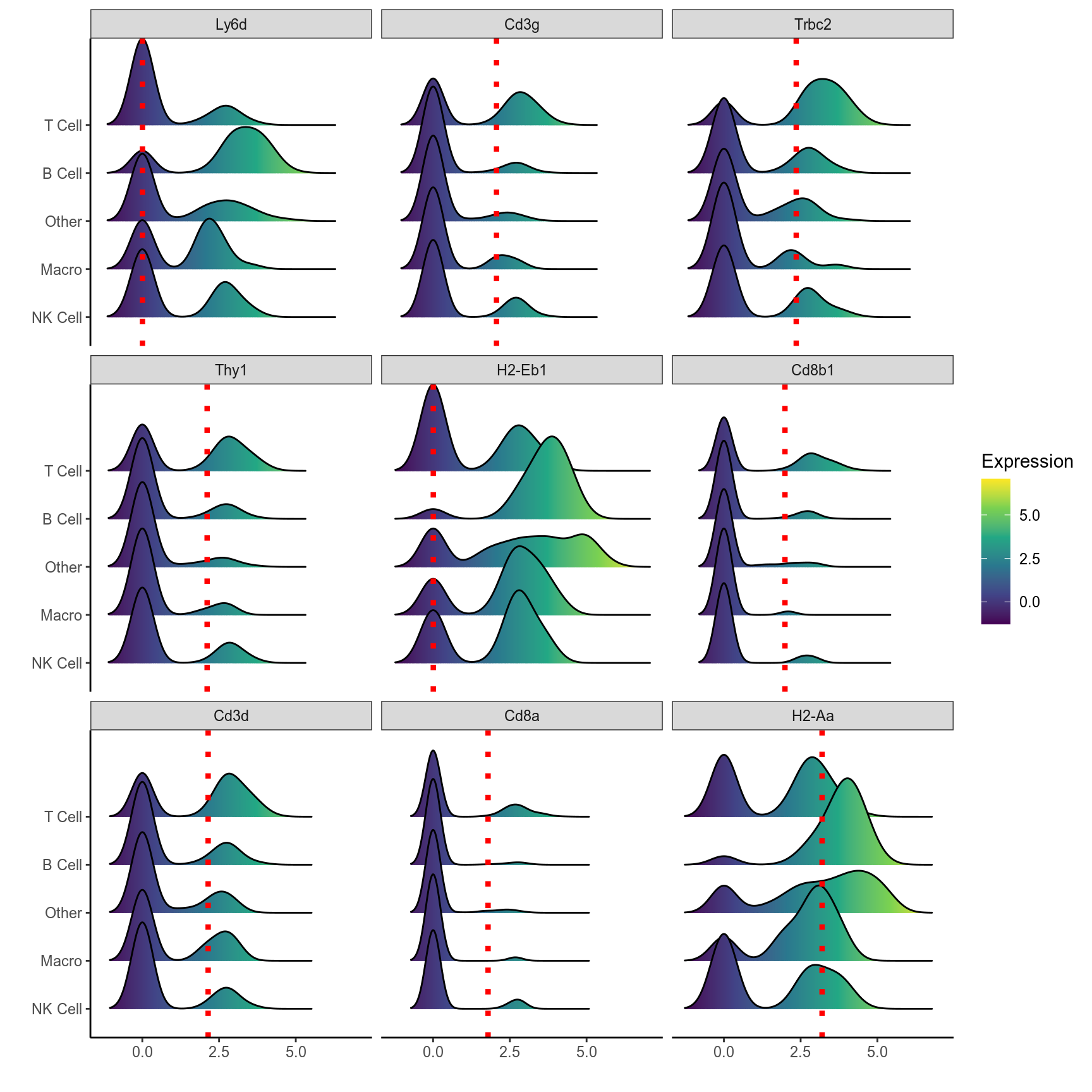
Markers identification for Macrophages
Macro.markers <- Detect_single_marker(mca.spleen, id = "Macro", category = "Flow", org = "mouse", min.pct = 0.1)
get_antibody(Macro.markers, org = "mouse")
# Ridge plot of top 9 markers of B cells
plot_ridge(mca.spleen, id = "Macro", genes = Macro.markers[1:9,]$gene, ncol = 3, assay = "RNA", aggr.other = F)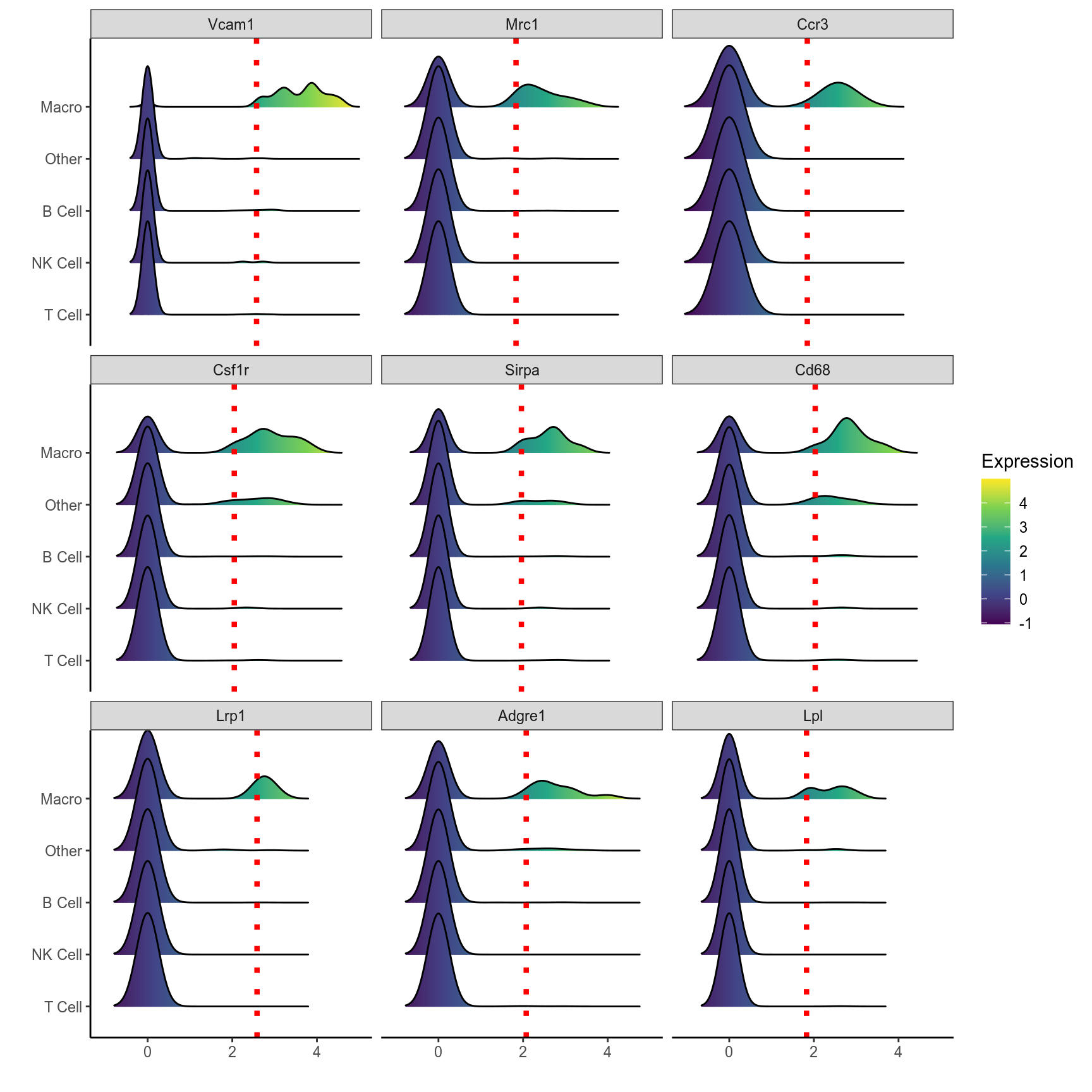
Session Info
sessionInfo()
#> R version 4.0.3 (2020-10-10)
#> Platform: x86_64-conda-linux-gnu (64-bit)
#> Running under: Linux Mint 18.3
#>
#> Matrix products: default
#> BLAS/LAPACK: /home/ronghui/anaconda3/envs/r4-real/lib/libopenblasp-r0.3.12.so
#>
#> locale:
#> [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
#> [3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
#> [5] LC_MONETARY=de_DE.UTF-8 LC_MESSAGES=en_US.UTF-8
#> [7] LC_PAPER=de_DE.UTF-8 LC_NAME=C
#> [9] LC_ADDRESS=C LC_TELEPHONE=C
#> [11] LC_MEASUREMENT=de_DE.UTF-8 LC_IDENTIFICATION=C
#>
#> attached base packages:
#> [1] stats graphics grDevices utils datasets methods base
#>
#> other attached packages:
#> [1] ggplot2_3.3.3 SeuratObject_4.0.0 sc2marker_1.0.3
#>
#> loaded via a namespace (and not attached):
#> [1] Seurat_4.0.1 Rtsne_0.15 colorspace_2.0-0
#> [4] deldir_0.2-10 ellipsis_0.3.2 ggridges_0.5.3
#> [7] rprojroot_2.0.2 fs_1.5.0 rstudioapi_0.13
#> [10] spatstat.data_2.1-0 farver_2.1.0 leiden_0.3.7
#> [13] listenv_0.8.0 DT_0.17 ggrepel_0.9.1
#> [16] fansi_0.4.2 codetools_0.2-18 splines_4.0.3
#> [19] cachem_1.0.4 knitr_1.31 polyclip_1.10-0
#> [22] jsonlite_1.7.2 ica_1.0-2 cluster_2.1.0
#> [25] png_0.1-7 uwot_0.1.10 shiny_1.6.0
#> [28] sctransform_0.3.2 spatstat.sparse_2.0-0 compiler_4.0.3
#> [31] httr_1.4.2 assertthat_0.2.1 Matrix_1.3-2
#> [34] fastmap_1.1.0 lazyeval_0.2.2 cli_3.3.0
#> [37] later_1.2.0 htmltools_0.5.2 tools_4.0.3
#> [40] igraph_1.3.0 gtable_0.3.0 glue_1.6.2
#> [43] RANN_2.6.1 reshape2_1.4.4 dplyr_1.0.7
#> [46] Rcpp_1.0.7 scattermore_0.7 jquerylib_0.1.3
#> [49] pkgdown_2.0.3 vctrs_0.3.8 nlme_3.1-152
#> [52] crosstalk_1.1.1 lmtest_0.9-38 xfun_0.30
#> [55] stringr_1.4.0 globals_0.14.0 mime_0.10
#> [58] miniUI_0.1.1.1 lifecycle_1.0.0 irlba_2.3.3
#> [61] goftest_1.2-2 future_1.21.0 MASS_7.3-53
#> [64] zoo_1.8-9 scales_1.1.1 spatstat.core_2.1-2
#> [67] promises_1.2.0.1 spatstat.utils_2.1-0 parallel_4.0.3
#> [70] RColorBrewer_1.1-2 yaml_2.2.1 memoise_2.0.0
#> [73] reticulate_1.19 pbapply_1.4-3 gridExtra_2.3
#> [76] sass_0.4.1 rpart_4.1-15 stringi_1.5.3
#> [79] highr_0.8 desc_1.2.0 rlang_1.0.2
#> [82] pkgconfig_2.0.3 matrixStats_0.58.0 evaluate_0.14
#> [85] lattice_0.20-41 ROCR_1.0-11 purrr_0.3.4
#> [88] tensor_1.5 labeling_0.4.2 patchwork_1.1.1
#> [91] htmlwidgets_1.5.3 cowplot_1.1.1 tidyselect_1.1.0
#> [94] parallelly_1.24.0 RcppAnnoy_0.0.18 plyr_1.8.6
#> [97] magrittr_2.0.1 R6_2.5.0 generics_0.1.0
#> [100] DBI_1.1.1 withr_2.5.0 mgcv_1.8-33
#> [103] pillar_1.6.4 fitdistrplus_1.1-3 survival_3.2-7
#> [106] abind_1.4-5 tibble_3.1.6 future.apply_1.7.0
#> [109] crayon_1.4.1 KernSmooth_2.23-18 utf8_1.2.1
#> [112] spatstat.geom_2.1-0 plotly_4.9.3 rmarkdown_2.14
#> [115] grid_4.0.3 data.table_1.14.0 digest_0.6.27
#> [118] xtable_1.8-4 tidyr_1.1.3 httpuv_1.6.0
#> [121] munsell_0.5.0 viridisLite_0.4.0 bslib_0.3.1