James S. Nagai1, Nils B. Leimkühler2, Michael T. Schaub 3, Rebekka K. Schneider4,5,6, Ivan G. Costa1*
1Institute for Computational Genomics, Faculty of Medicine, RWTH Aachen University, Aachen, 52074 Germany
2Department of Hematology and Stem Cell Transplantation, University Hospital Essen, Germany
3Department of Computer Science, RWTH Aachen University, Germany
4Department of Cell Biology, Institute for Biomedical Engineering, Faculty of Medicine,RWTH Aachen University, Pauwelsstrasse 30, 52074 Aachen, NRW, Germany
5Oncode Institute, Erasmus Medical Center, Rotterdam, 3015GD, the Netherlands
6Department of Hematology, Erasmus Medical Center, Rotterdam, 3015GD, the Netherlands
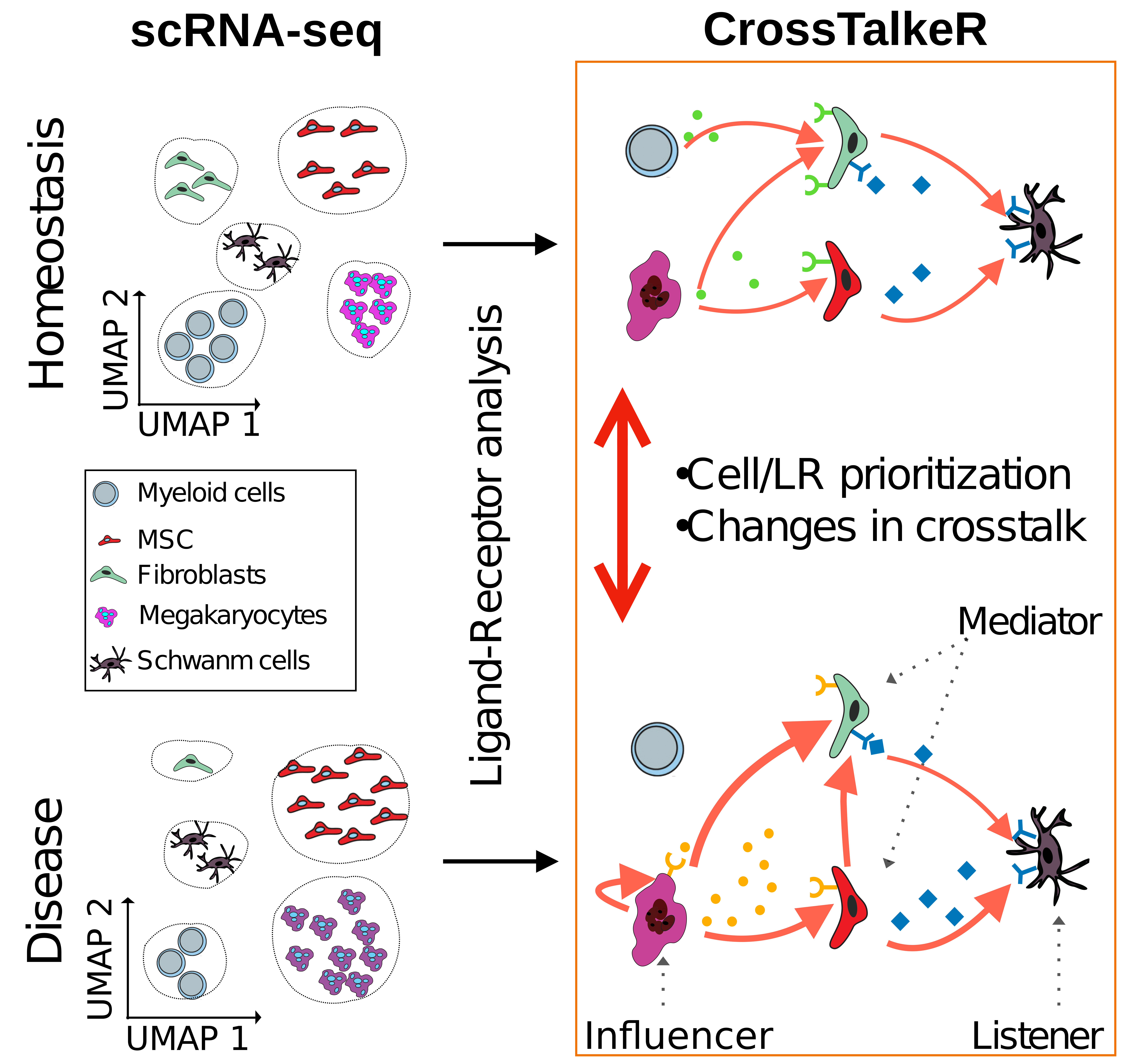
Motivation: Ligand-receptor (LR) analysis allows the characterization of cellular crosstalk from single cell RNA-seq data. However, current LR methods provide limited approaches for prioritization of cell types, ligands or receptors or characterizing changes in crosstalk between two biological conditions.
Results: CrossTalkeR is a framework for network analysis and visualisation of LR networks. CrossTalkeR identifies relevant ligands, receptors and cell types contributing to changes in cell communication when contrasting two biological states: disease vs. homeostasis. A case study on scRNA-seq of human myeloproliferative neoplasms reinforces the strengths of CrossTalkeR for characterisation of changes in cellular crosstalk in disease state.
Install
You can install CrossTalkeR with the simple commands below:
install.packages("devtools")
devtools::install_github("https://github.com/CostaLab/CrossTalkeR", build_vignettes = TRUE)
require(CrossTalkeR)Note: Please avoid to use the following characters in celltype name: ‘$’
Possible system dependencies
libudunits2-dev
libgdal-dev
gdal-bin
libproj-dev
proj-data
proj-bin
libgeos-devCrossTalkeR Plots examples and vignette
We provide in our vignette examples on how to analyse cell interactions from a human myelofibrosis single cell RNA-seq.
vignette('CrossTalkeR-HumanMyfib')CrossTalkeR Python Package 🐍
Our package is now available in Python — bring differential cell-cell communication analysis to your Python environment! You can find more information in our Read the Docs
CrossTalkeR Docker image
We provide access to a Docker image, available at: https://gitlab.com/sysbiobig/ismb-eccb-2025-tutorial-vt3/container_registry. The Docker image comes preconfigured with all necessary libraries, tools, and software required to follow the hands-on exercises.
🔥 CrossTalkeR Realease v2.0 - New Features 🔥
Statistical Filtering with Fisher’s Exact Test
Filter cell-cell communication networks using Fisher’s test to identify statistically significant interactions.Volcano Plot Visualization
Visualize results of the Fisher’s test in a volcano plotHeatmap Visualization with Clustering
Explore communication patterns across cell types using heatmaps, including clustering by interaction weights-
Comprehensive Topological Analysis
Generate bar plots for the calculated network topological measures, separately for:- cell–cell interaction networks
- cell-gene interaction networks
- cell–cell interaction networks
Ligand–Receptor Enrichment Analysis with PROGENy
Step-by-step tutorial for pathway enrichment analysis of ligand–receptor pairs using PROGENy.Modeling Intracellular Communication
Extend the communication network by incorporating transcription factors to model intracellular signaling.Integration with LIANA+
Seamlessly using ligand-receptor interaction results from LIANA+. We provide a detailed tutorial on how to perform the integration Run lianaCompatibility with scSeqComm
UsescSeqCommoutputs as inputs to the CrossTalkeR framework for downstream comparative analysis.
Features v1.4.0
- Splitted generate_report function in two parts:
- analise_LR() to only run the analysis without generating the CrossTalkeR report
- make_report() to only generate a new CrossTalkeR report for existing CrossTalkeR results
- Added node types to the network:
- we now consider the annotation of a gene as ligand (L) or receptor (R) to consider the biological function
- Less constrains on the cell cluster name annotation (only ‘$’ must be avoided in the cluster naming)
- Integration with liana-py for ligand-receptor interaction predictions
Features v1.3.0
- Single and Comparative Reports
- Cell Cell Interaction visualization
- Sending and Receiving Cells Ranking
- Gene Target based Sankey Plots
- CCI and GCI PCA ranking
- All measures and PC table
- PC1 and PC2 based barplot
- Leimkühler et. al. [2] data were added to the package
- Fisher Test were implemented to highlight the CCI edges significance (new)
-
Change input format: Please see the Documentation
- A python3 notebook are available to cast the old input to the new input.
- Liana (Dimitrov et. al. [3]) Output can be used as CrossTalkeR input.
- LR pair visualization plot can be done using a Seurat Object



